
| Species | Virus | Abbreviation | |
| Antheraea pernyi iflavirus | Antheraea pernyi iflavirus | APV | |
| Brevicoryne brassicae virus | Brevicoryne brassicae virus | ||
| Deformed wing virus | deformed wing virus | DWV | |
| Kakugo virus | KV | ||
| Dinocampus coccinellae paralysis virus | Dinocampus coccinellae paralysis virus | ||
| Ectropis obliqua virus | Ectropis obliqua virus | EoV | |
| Infectious flacherie virus* | infectious flacherie virus | IFV | |
| Lygus lineolaris virus 1 | Lygus lineolaris virus 1 | LlV-1 | |
| Lymantria dispar iflavirus 1 | Lymantria dispar iflavirus 1 | ||
| Nilaparvata lugens honeydew virus 1 | Nilaparvata lugens honeydew virus 1 | NlHV-1 | |
| Perina nuda virus | Perina nuda virus | PnV | |
| Sacbrood virus | sacbrood virus | SBV | |
| Slow bee paralysis virus | slow bee paralysis virus | SBPV | |
| Spodoptera exigua iflavirus 1 | Spodoptera exigua iflavirus 1 | ||
| Spodoptera exigua iflavirus 2 | Spodoptera exigua iflavirus 2 | ||
| Varroa destructor virus-1 | Varroa destructor virus 1 | VDV-1 | |
| *, type species |
It is clear from sequence comparisons (N.J. Knowles, unpublished observations) that 10 of these viruses can be divided into seven groups (species or genera?) which I have arbitrarily labelled A to G:
A) Infectious flacherie virus (IFV)
B) Sacbrood virus (SBV)
C) Perina nuda virus (PnV) and Evtropis oblique picorna-like virus (EoPV)
D) Deformed wing virus (DWV), Varroa destructor virus 1 (VDV-1) and Kakugo virus (KV)
E) Venturia canescens picorna-like virus (VcPLV)
F) Brevicoryne brassicae picorna-like virus (BBPLV)
G) (Honey) slow bee paralysis virus (BSPV)
Below is a N-J tree of
the polymerase region (~500 aa). Each of the seven virus groups could
represent a distinct genus.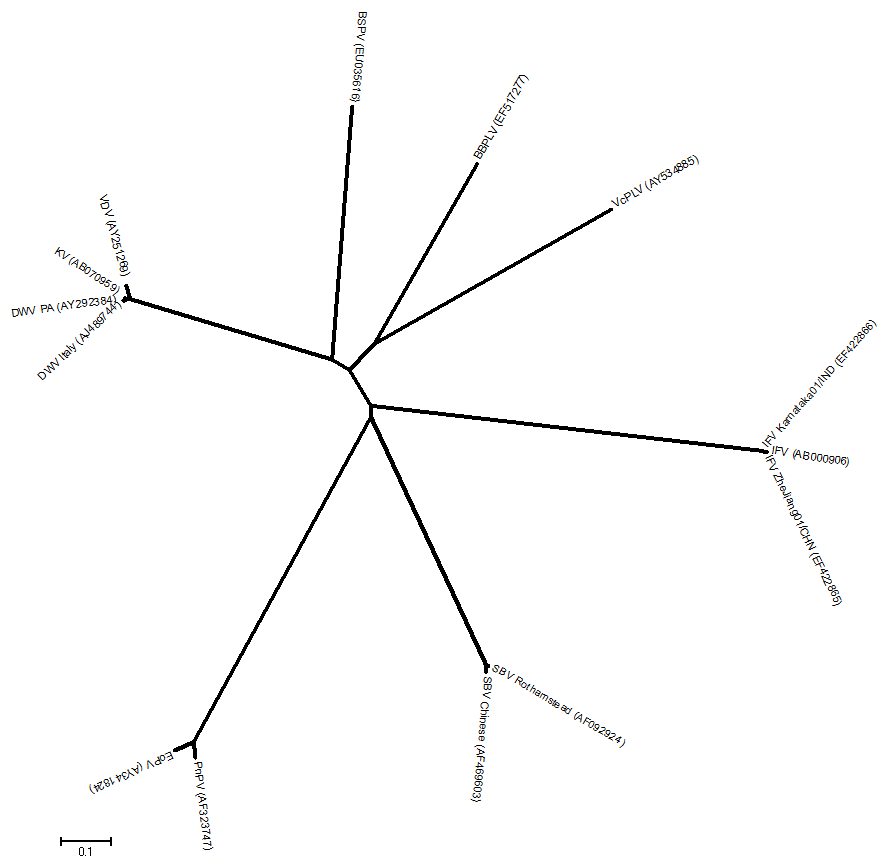
Software: MEGA 3.1
No. of Taxa : 14
Data File : Iflaviridae_pol_2.meg
Data Title : Iflavirus partial pol
Data Type : Amino acid
Analysis : Phylogeny reconstruction
Tree Inference : ==============================
Method : Neighbor-Joining
Phylogeny Test and options : Bootstrap (1000 replicates; seed=64238)
Include Sites : ==============================
Gaps/Missing Data : Pairwise Deletion
Substitution Model : ==============================
Model : Amino: Poisson correction
Substitutions to Include : All
Pattern among Lineages : Same (Homogeneous)
Rates among sites : Uniform rates
No. of Sites : 544
No Of Bootstrap Reps = 1000
Table showing the percentage amino acid differences for the polymerase region depicted in the tree above.
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | 13 | 14 | ||
| 1 | IFV (AB000906) | 0 | |||||||||||||
| 2 | IFV (EF422865) | 0.8 | 0 | ||||||||||||
| 3 | IFV (EF422866) | 0.8 | 0.4 | 0 | |||||||||||
| 4 | SBV (AF092924) | 74.5 | 74.5 | 74.5 | 0 | ||||||||||
| 5 | SBV (AF469603) | 74.9 | 74.9 | 74.9 | 1.7 | 0 | |||||||||
| 6 | PnPV (AF323747) | 78.8 | 78.8 | 78.8 | 73.6 | 73.8 | 0 | ||||||||
| 7 | EoPV (AY341824) | 79.2 | 78.8 | 78.8 | 72.7 | 72.9 | 6.9 | 0 | |||||||
| 8 | DWV (AJ489744) | 75.1 | 75.1 | 74.8 | 66.3 | 66.7 | 73.9 | 74.3 | 0 | ||||||
| 9 | DWV (AY292384) | 75.2 | 75.2 | 74.9 | 66.5 | 66.9 | 74 | 74.4 | 0 | 0 | |||||
| 10 | VDV (AY251269) | 75.4 | 75.4 | 75.2 | 66.9 | 67.3 | 74.6 | 75.5 | 3.9 | 3.9 | 0 | ||||
| 11 | KV (AB070959) | 75.1 | 75.1 | 74.9 | 66.2 | 66.6 | 74.2 | 74.4 | 1.2 | 1.2 | 3.9 | 0 | |||
| 12 | BBPLV (EF517277) | 74.9 | 74.7 | 74.7 | 65.9 | 65.7 | 75.4 | 76.2 | 58.7 | 58.7 | 59.3 | 58.8 | 0 | ||
| 13 | VcPLV (AY534885) | 76.2 | 75.8 | 76 | 71.1 | 71.1 | 77.9 | 78.3 | 65.4 | 65.6 | 65.6 | 65.1 | 61.2 | 0 | |
| 14 | BSPV (EU035616) | 74.9 | 74.7 | 74.5 | 70.8 | 71.2 | 74.1 | 74.7 | 60.8 | 60.7 | 61.1 | 60.5 | 65.2 | 70.1 | 0 |